An interview with Ingmar Riedel-Kruse and Paulo Blikstein about their NSF-funded project to build and evaluate a technological and curricular infrastructure to empower scalable, low-cost experimentations for undergraduates and K-12 students in the life sciences, combining them with computer models in real time.
What is the big idea of your project?
The big picture is: How can we make microbiology interactive and available to everyone? And how can we combine experimentation with computer modeling in real time? How can all ages, from child to adult, experience what microscopic life is like? Right now what we have in schools is microscopy, where students passively observe. It’s hard to do experiments and for teachers to bring in biological organisms––harder than physics, for example, where you can put something back in a drawer at the end of the class.
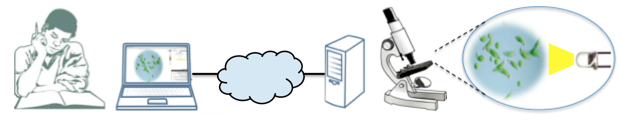
So what we have been developing is different types of platforms to make easy two-way interaction between humans and microscopic organisms possible. We also believe that sensemaking is more powerful when experiments are tightly connected with models and simulations, instead of being separate activities—thus we are using the idea of Bifocal Modeling to combine our biological experiments and models in real time. For example, we have an online platform where you can remotely observe Euglena cells swimming in a small dish by streaming video. You can shine light on the small organisms using a virtual joystick control, and when the light hits the cells, you can see how they swim away, in real time. We use two cameras: one for the internal view into microscope where you can see the cells, and the other is the external “macroscopic” view where the student can see the light source. At the same time, we have a modeling environment that enables students to program a model to mimic what they just saw in the real world. By trying to “match” the real world, and realizing the differences between real and ideal systems, students dive deeper into the Euglena’s behaviors. This is all fully automated, almost care-free on our end. For a couple of days to weeks, the system just works on its own. The physical lab is 10 meters from where we are sitting, and anyone can join the lab online.
Who has used your interactive labs, and how have they used it?
We did tests with undergrad and graduate students initially, and a mini online course for 1-2 hours where students could do some simple experiments. We recently completed our first testing with about 100 middle school students across 8 groups in San Francisco. We’re also talking with schools in Oakland.
The primary experiment, as I explained, is that you see cells, you put light stimulus on them, you see a response, and then the system actually collects the data for you. It saves both the stimulus that you did as well as the images of how the cells reacted. Then you can go to another user interface where you can replay your data, and start analyzing your experiments––count how many cells responded, how fast they moved, and so on. We also have made a computer modeling environment where you can try to replicate the behavior of the real organism and change a few parameters––like how fast does the cell move, how strongly does it respond to light. You can do your experiment and then do your modeling, and compare the two side by side.
So our interactive biotechnology is enabling students and teachers to do some very convenient free-play experimentation that can eventually include all parts of science because you can explore, do guided experiments, make models, generate hypotheses (like “does the cell move faster when the light is on or off”), and make measurements to test your hypotheses. By having this all online, it takes a lot of the load off of the teacher because students can do experiments anytime, even in the evening. We see students logging in at crazy hours to see what is going on. They are really hooked! In the beginning, they are very playful, doing fun experiments. Over time, this playfulness evolved into more principled testing. They transitioned into behaving more like scientists. The technology enables it. Scientists also play and find things by chance; they don’t know, they have crazy ideas, and there’s a lot of creativity and exploration, especially in the early phases. Then they move into more rigorous testing. So one important aspect of the system is that is allows student to explore scientific discovery differently than what’s mandated in the “traditional” version of the scientific method. They can start playing around and overtime become more methodical—and the fact that they can interact with the experiments for several hours, anywhere and anytime, has a lot to do with it.
We’re also incorporating learning analytics to track what the students are doing and to learn about what they are learning, and have done interviews and recorded video of students engaging with the system so we can see how they behave. We also have other systems with different types of organisms and experiments that occur over longer timescales, and a maker project, which we demonstrated at Cyberlearning 2015, where children build a little microscope, attach a phone to the microscope to observe similar organisms, and the phone does the data processing and analysis.
Where do you see your work going in the future?
We see this as new technology that should really enable millions of kids in the future to do biology experiments in formal and informal settings, and a new window into learning research.
More generally, the distance between what kids do in science labs in schools and what is done in real science labs is greater than it has ever been. There are a lot of new things that scientists now have at their disposal and are using in their research, like computational models, automated data collection, image processing that can interpret results, and so on. A lot of new things that everyone is using. But kids still do the same stuff in school. They float bacteria in a petri dish and look at them for a couple of days. There are a lot of classic experiments that are like legacy there. But they are less and less interesting because there are lots of more interesting problems the world is trying to deal with. Even with the Euglena, there are a lot of real-world applications around contamination and pollution. There are a lot of scientific problems closer to real life, but kids don’t have access to them because they are still stuck in 19th century style experiments.
The problem is that to bring more interesting experiments to schools is hard because you need a big infrastructure and the technical knowledge about how to maintain it, if that’s even possible. Labs are very expensive. And we don’t want to take 200 year-old experiments and put them online in a model. We don’t believe that simply taking the outdated stuff we do in schools and packaging it in colorful simulations will do it. Our approach is to put today’s experiments online––not just to cut costs, but because they are more interesting and would be hard to do in real schools because they are technically very complicated. Cutting costs is good, but sometimes it’s not the main point. Let’s bring the most interesting and complex stuff and make it feasible in schools. Let’s stop thinking of “online” as a way to simply make education massive and cheap, but to make it more interesting, more complex, and enable children to learn things that were unimaginable before.
NSF Project Information
Title: DIP: Collaborative Research: Taking Hands-on Experimentation to the Cloud: Comparing Physical and Virtual Models in Biology on a Massive Scale
Award Details
Investigators: Ingmar (Hans) Riedel-Kruse, Paulo Blikstein
Also, there is a reason scientists do experiments: it’s not just because they already know the stuff and they need to just run experiments. They are useful to brainstorm new ideas to find out new things. So we also try to keep what is rich about the experiment in this system, not sanitize it too much so that it doesn’t look real. In real life, you never get a perfect result. When you move the joystick to move the light, not all of the Euglena go away from the light. Some just stay there and go in circles, some even go towards the light. We talk to students and ask, “Why is this happening?” Instead of avoiding that confusion, we engage students in conversation about that. They understand that maybe some of them are very old, they aren’t reactive anymore, maybe their sensor is broken, their eye doesn’t work any more, maybe they are starving; there is some natural variability, just like with people. Some people run faster, some are blind, and so on. The cells aren’t all exactly the same, and that is very generative. The notion that experiments have noise and are inexact is not something you should hide from students. You should actually make kids talk about it. They have very rich discussions, and that is really the spirit of the idea of Bifocal Modeling.
How do you work with teachers?
In the local schools, we tested a 1 hour 50 minute curriculum with sequence of student activities, questions, and pre- and post- test. We did a lot of co-design of the curriculum with teachers, and are refining them so teachers can easily enact them in a normal classroom. For this unit, we worked with teachers for about 5-8 weeks, showing them what we’ve created, and asking them what’s feasible.
In Paulo’s CAREER grant on Bifocal Modeling, we have also been codesigning units with teachers. We went to the school and talked with teachers about what topics they were covering, and a graduate student met with a teacher every week. They went through the topics she covered, picked a topic, and together codesigned a unit on polar and apolar molecules. The teacher picked an oil spill as a theme for the unit, around cleaning with detergents. The teacher was very much an author of the unit, and we learned a lot. We are also working toward making it easier for teachers to create and adapt content on their own in our system. Sometimes teachers will try things because they are a novelty, but if you don’t have them codesign and feel ownership, they’re not going to use it once you leave.
We try to take a layered approach where we start by working with one teacher, then bring in others and have them work together; the teacher who did the codesign can coach the new teacher. So in a school you create a small community of teachers who are well-versed. We try to get them to document what they do in a language that other teachers would understand. Eventually you try to run a professional development program where you bring in other teachers over a weekend. Hopefully at some point have a critical mass of people and materials that become part of professional development programs.
Are there things that CIRCL could be doing to help your work with teachers?
At our FabLearn conference, the participants are roughly half teachers and half researchers. Some teachers haven’t seen a researcher in their life! We have researchers and teachers presenting, and I think that helps teachers internalize the research language, and also helps the researchers. I think ideally it’s great to have events where researchers and teachers co-mingle and both present to each other.
Can you say more about the cyberinfrastructure for this work?
Generally, bioengineering technologies are getting better and cheaper in terms of how much biology they can manipulate and measure, how they scale, and how they become higher throughput, so will become easier and easier to build systems and connect them to schools––similar to what happened with the electronics revolution and transistors and electronic circuits. The devices we have are most often used for diagnostics, and many things are being automated, so a student could use them to program and run their own experiments. One experiment might take a minute. And we have an asynchronous mode where you can program something and submit it to a queue to execute. So you could in principle do a thousand experiments a day on one machine. You can see how 10-20 subsystems, managed well, could service a substantial population across the world.
We have a very interdisciplinary group of graduate students in computer science, bioengineering, physics, and education working together to design and build these things. We make our code and building plans available in GitHub so that others can recreate them. The initial technology is just a good microscope; the basic unit should be cheap, between $100-$500. Then you hook it up to the web. We have 2 webcams, and external one and internal one, and an Arduino is actuating LEDs. If you just take the microscope away and put something else in there, controlled, say, by an Arduino, this could be a physics experiment. For example, you could put in a LEGO robot. We have a teacher coming to work with us over the summer on LEGO robots, so that kids in robotics classes could make robots like the ones we use to run our experiments but in a kind of mini-cloud, not even necessarily online. So this whole platform could be used in different ways for doing online experiments. All the hardware is off the shelf and the software is open source. We also have a mini server in the grant that can run in the school. But then the students can’t use it at night or as easily run week-long experiments. For some things it might make sense to run the experiment at a university or company. But schools could run their own lab. There should be multiple infrastructure models.
In the end, it’s not just about putting labs online. It’s about labs with persistence, so you can go back to them, experiments that can be a week long, you can track what is happening. It’s a different type of hands-on science that keeps the richness and authenticity of real science labs.
For more information
Biotic Games-Playing with Living Cells (YouTube Video of a talk)
Interactive Cloud Experimentation for Biology: An Online Education Case Study. Describes a cloud architecture for sharing and executing biology experiments in parallel, remotely and interactively, to lower the access barrier for non-biologists, enable discovery, and facilitate learning analytics.